Lecture 11: Cross Validation
DATA 101: Making Prediction with Data
University of British Columbia Okanagan
Introduction
- In the last lecture we saw how we could fit the KNN classification models using tidymodels.
- Recall: tidymodels is an R package and framework for modeling that follows the principles of the tidyverse.
- In the upcoming lectures we will continue to work with tidymodels for evaluation and tuning
tidymodels packages
 recipes: tidy interface for data pre-processing
recipes: tidy interface for data pre-processing
 parnips: tidy interface for fitting models
parnips: tidy interface for fitting models
 rsample: tidy interface for data splitting and resampling
rsample: tidy interface for data splitting and resampling
 yardstick tidy interface for validating models
yardstick tidy interface for validating models
Why tidymodels?
- R is open source – made by different people and using different principles, everything has a slightly different interface, and trying to keep everything in line can be frustrating
- tidymodels offers a consistent interface with many other R packages who do the work of fitting the models
Steps of tidymodels
- Build and fit a model
- Preprocess your data with recipes
- Evaluate your model with resamples
- Tune model parameters
- A predictive modeling case study
Steps of tidymodels
- Preprocess your data with recipes
- Build and fit a model
- Evaluate your model with resamples
- Tune model parameters
- A predictive modeling case study
We’ll go through building the model first but typically the pre-processing happens before training your model
Cancer Data
Let’s continue with our example from last class:
Recall that this is a classification problem in which we wish to classify tumors as either being malignant or benign.
Scatterplot
Code
# create scatter plot of tumor cell concavity versus smoothness,
# labeling the points be diagnosis class
perim_concav <- cancer |>
ggplot(aes(x = Smoothness, y = Concavity, color = Class)) +
geom_point(alpha = 0.5) +
labs(color = "Diagnosis") +
scale_color_manual(values = c("orange2", "steelblue2")) +
theme(text = element_text(size = 12))
perim_concav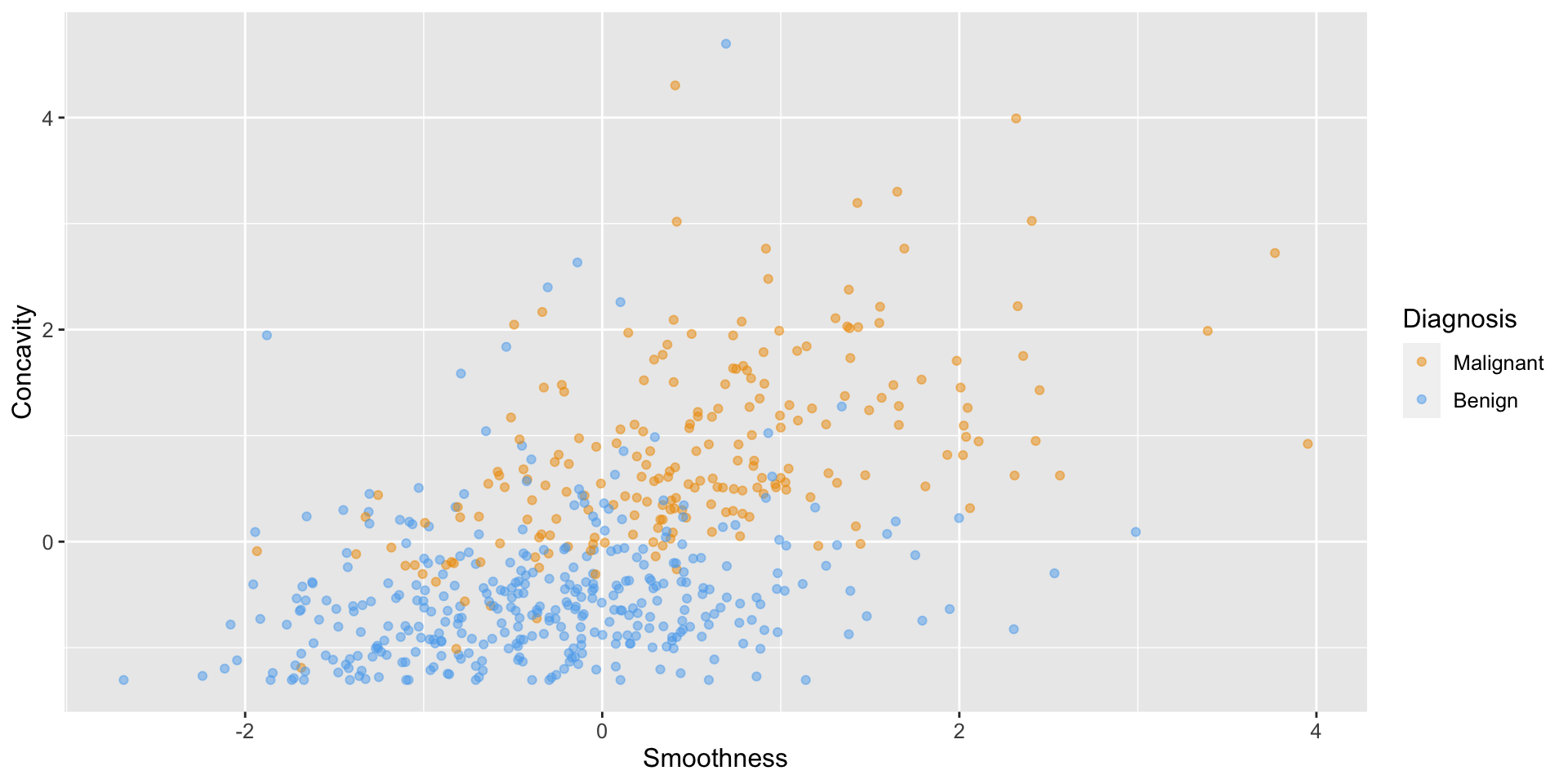
Create Training/Test set
Before we do anything, we want to split our data into a training and testing test…
1. Build a model
Parsnip categorizes models by:
Model type
To predict the label of a new observation (here, classify it as either benign or malignant), we will used the K-nearest neighbors (KNN) classification algorithm.
The model type is related to the structural aspect of the model. For example, the model type
linear_regrepresents linear models (slopes and intercepts) that model a numeric outcome. Other model types in the package arenearest_neighbor,decision_tree, and more.
Model engine
The computation engine is a combination of the estimation method and the implementation. For example, for linear regression, one engine is “lm” which uses ordinary least squares analysis via the lm() function. Another engine is “stan” which uses the Stan infrastructure to estimate parameters using Bayes rule.
Model mode
The mode is related to the modeling goal. Currently the two modes in the package are regression and classification. Some models have methods for both models (e.g. nearest neighbors) while others have only a single mode (e.g. logistic regression).
Fit the model
Now we train the classifier using the predictors Perimeter and Concavity using the fit() function
parsnip model object
Call:
kknn::train.kknn(formula = Class ~ Perimeter + Concavity, data = data, ks = min_rows(6, data, 5), kernel = ~"rectangular")
Type of response variable: nominal
Minimal misclassification: 0.07042254
Best kernel: rectangular
Best k: 6More than 2 predictors
If we wanted to use every variable in (exepect Class) as a predictor in the model, we could use a convenient shorthand syntax above which is the same as:
2. Pre-process
The recipes package which is designed to help you preprocess your data before training your model. Examples include:
- converting categorical predictors to indicator variables (also known as dummy variables),
- transforming data to be on a different scale (e.g., taking the logarithm of a variable),
- extracting key features from raw variables (e.g., getting the day of the week out of a date variable),
Normalizing
Remember in our discussion of KNN we said it was useful to normalize our data before fitting?
The above will normalize numeric data to have a standard deviation of one a mean of zero.
Now let’s refit the model
Workflow
Prediction
Now that we have a fitted KNN model, we can use it to predict the class labels for our test set.
Comment
To assess our model we will need to compare the true class of the patients in the test set (
cancer_test$Class) with the predict class we just outputted on the previous slide.To facilitate these comparison let’s save this information to new data frame
Evaluate performance
Finally, we can assess our classifier’s performance. First, we will examine accuracy. To do this we use the metrics function from tidymodels, specifying the truth and estimate arguments:
Confusion Matrix
We can also look at the confusion matrix using conf_mat:
Other metrics
conf_mat = confusion[[1]]
(accuracy <- sum(diag(conf_mat)) / sum(conf_mat))
(recall <- conf_mat[2, 2] / sum(conf_mat[2, ]))
(precision <- conf_mat[2, 2] / sum(conf_mat[, 2]))[1] 0.8461538
[1] 0.8695652
[1] 0.8888889Tuning Parameters
- The vast majority of predictive models in statistics and machine learning have tuning parameters.
- A tuning parameter (or hyperparamter) is a number you have to pick in advance that determines some aspect of how the model behaves.
- For the KNN classification algorithm,
K, the value that determines how many neighbors participate in the class vote, is a tuning parameter. - By picking different values of
K, we create different classifiers that make different predictions.
Tuning the Classifier
“Tuning a model”, refers to the process of adjusting the hyperparameters of a machine learning algorithm to optimize its performance on a specific task or dataset.
The goal of tuning a model is to find the best set of hyperparameter values that result in a model that generalizes well to unseen data, provides better accuracy1, and is well-suited to the specific problem at hand.
In our example tuning the model equates to how we pick the best value of
K
Unseen data
- The first step in choosing the parameter
Kis to be able to evaluate each classifier1 - The “best” classifier, would ideally, be the one which gets the accuracy on data it hasn’t seen yet.
- Problem: We cannot use our test data set in the process of building our model 🤔
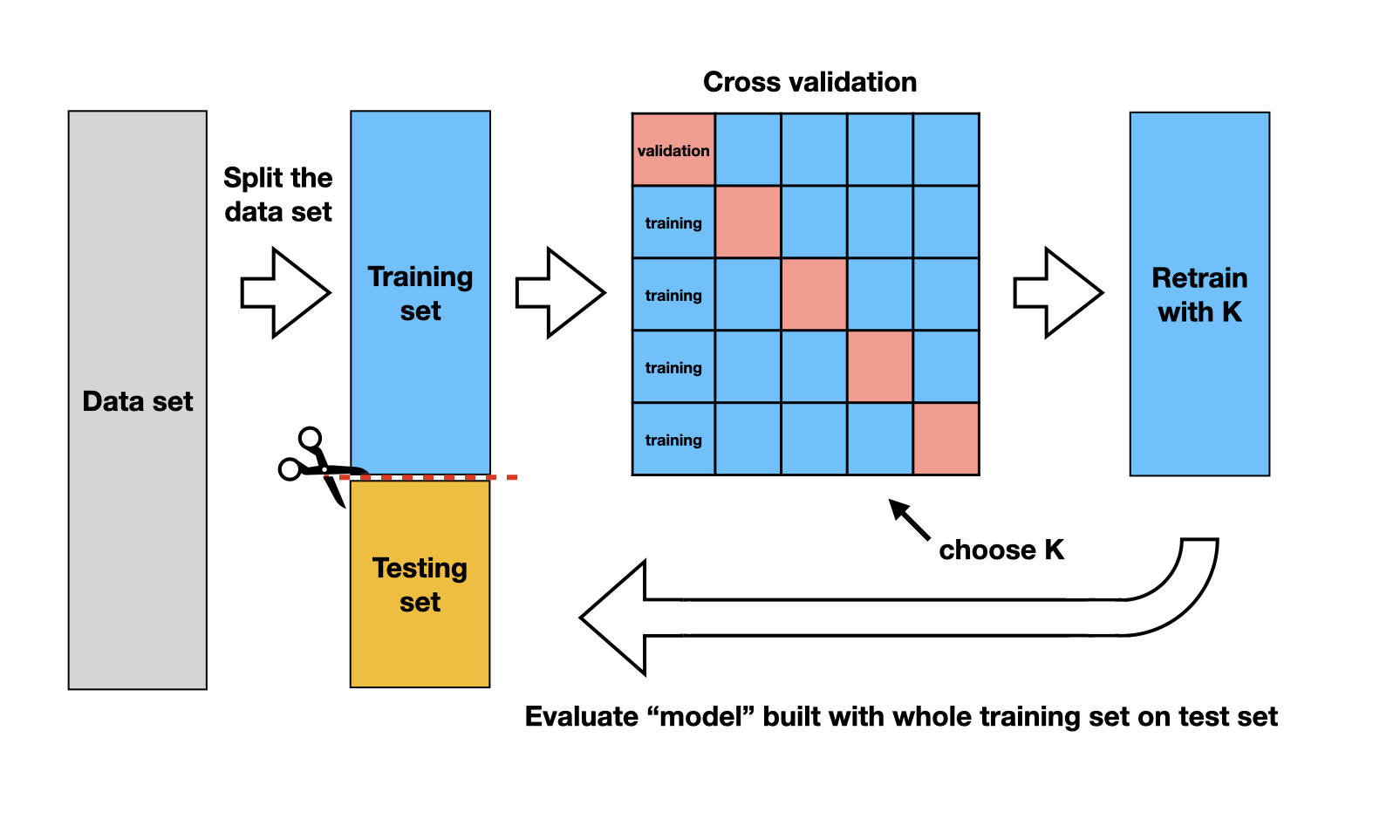
- Solution: Cross-validation!
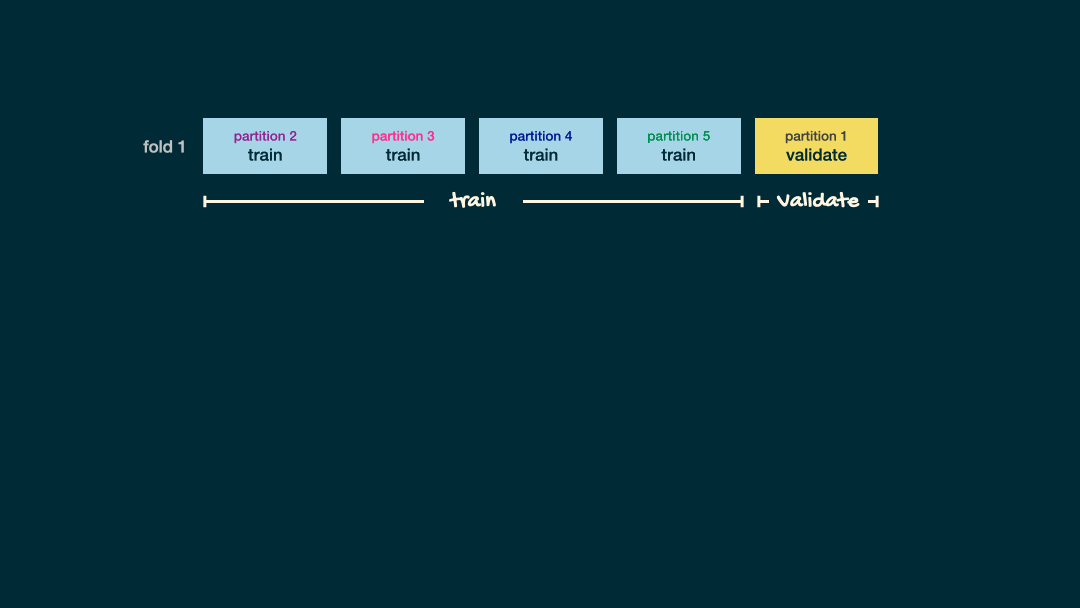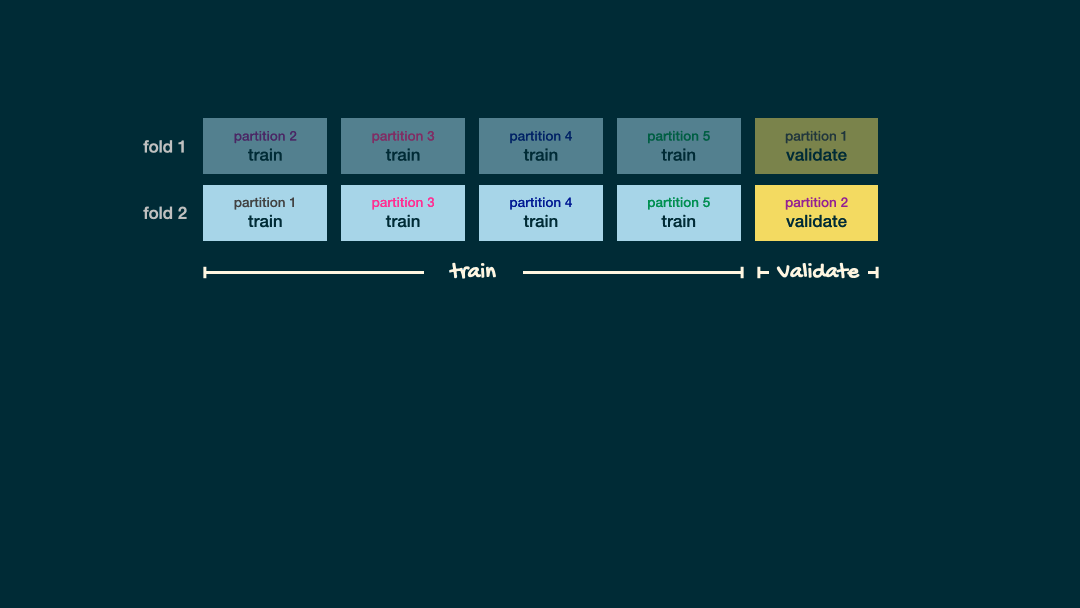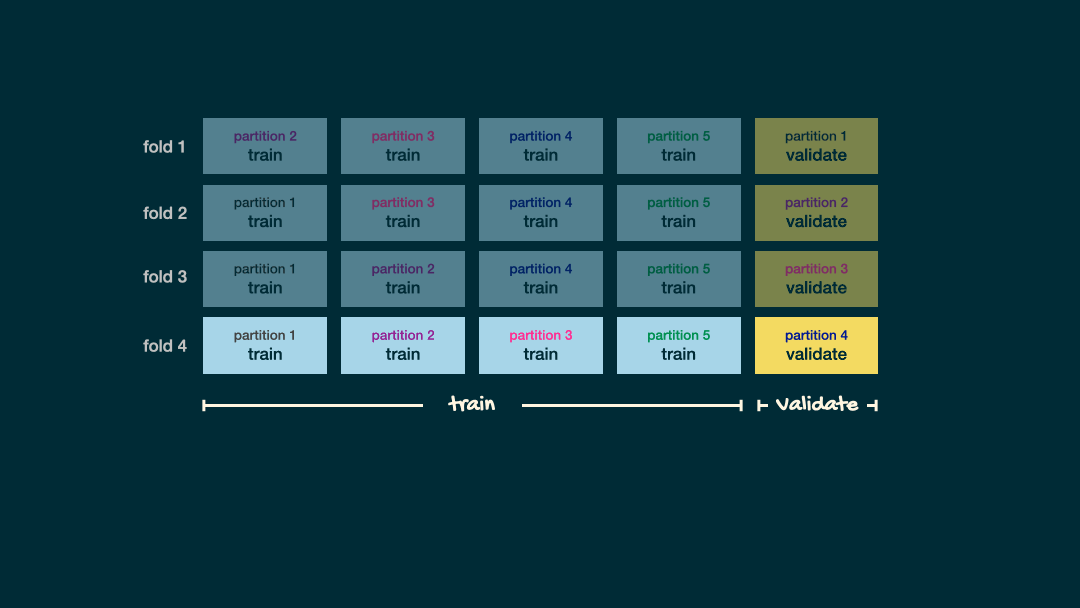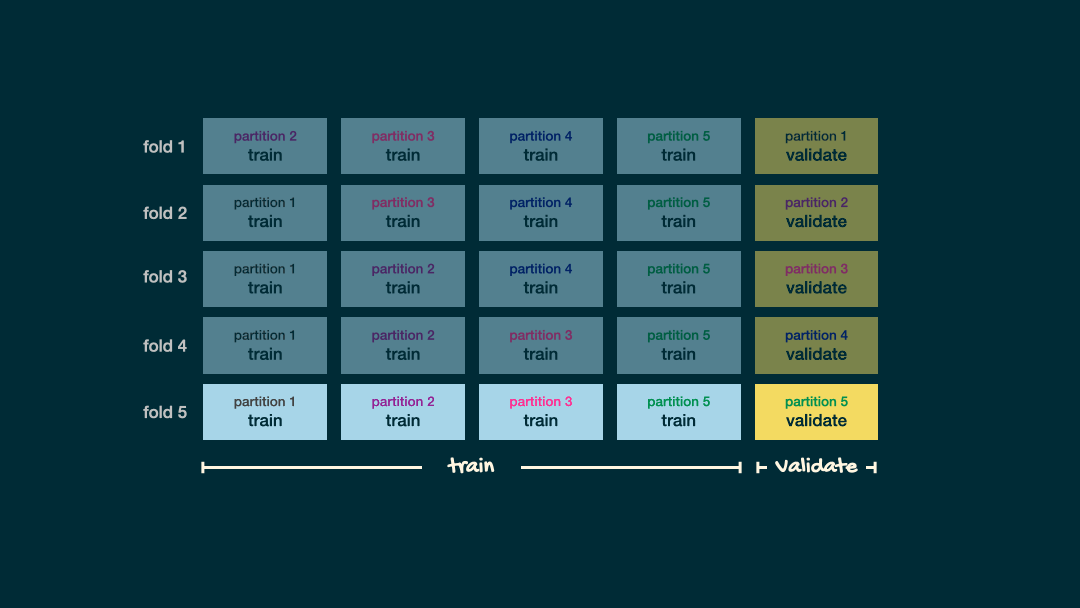
Cross-Validation
V-fold Cross-validation (CV) uses the same trick did before when evaluating our classifier, that is we will “hide” some data during the fitting process
- For example, 5-fold CV involves splitting the training data into five subsets
- Each subset will have a turn as the validation set
- That is, we will have 5 validation sets evaluating the five different fits (based five different subsets).
- We then aggregating the results to estimate how well the model is perform for that value of the hyperparameter
Cross-validation schematic
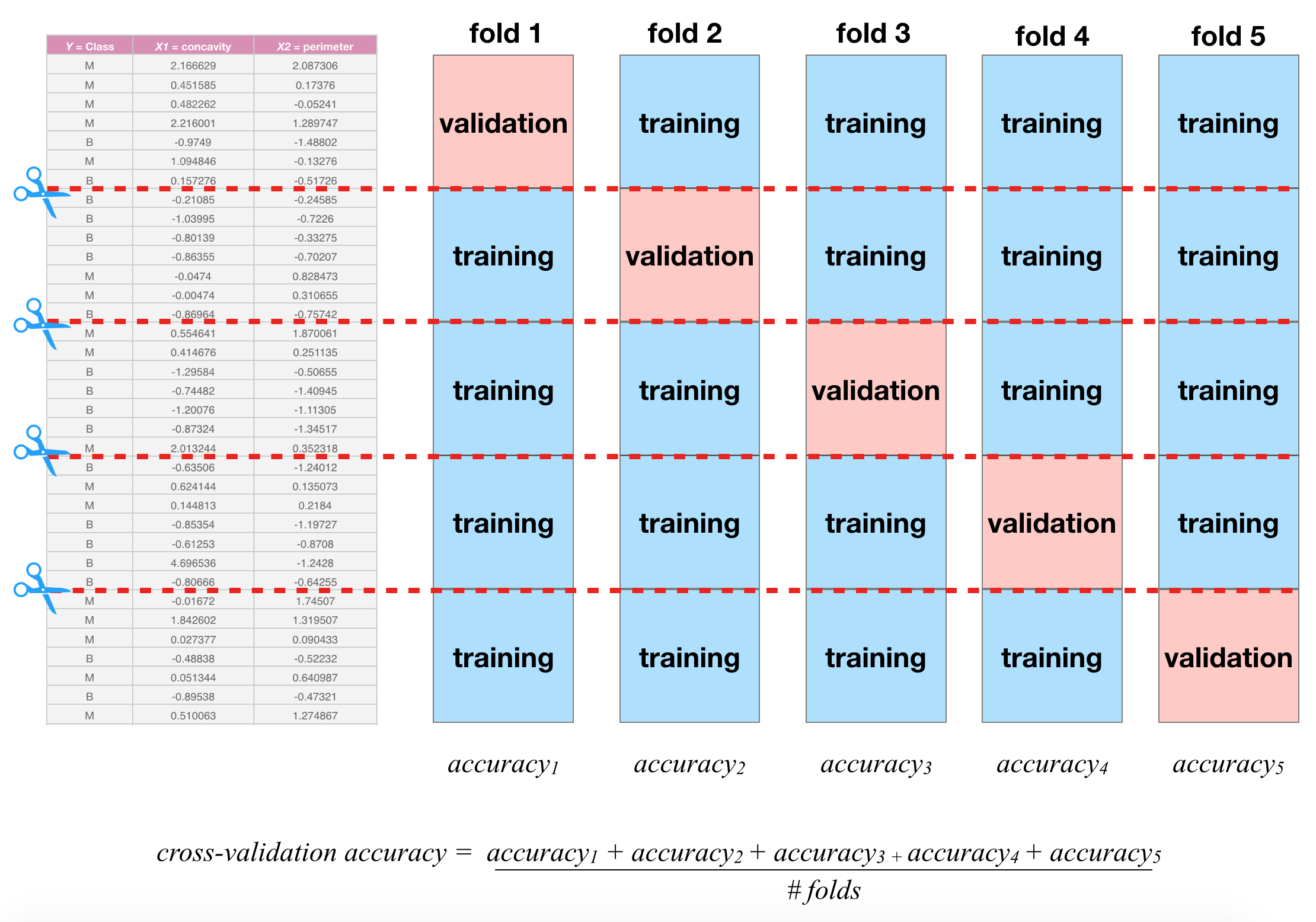
Depiction of 5-fold cross validation. Note that the data would be shuffled before creating these folds. Source Sec 6.6.1
Steps for tuning a model
- Define a range or a set of possible values for the hyperparameter(s).
- Select an evaluation metric that quantifies the performance of the model (e.g. accuracy)
- Use cross-validation to estimate the model’s performance with different hyperparameter combinations.
- Select the hyperparameter value(s) that result in the best model performance according to the chosen evaluation metric.
Visual example
For each of your potentail values of your hyperparameter you fit V models and calculated the CV-accuracy


\(\dots\)



Visual example
The model which produces the best CV-accuracy is deemed the “best” model.



\(\dots\)



CV in R
To perform 5-fold CV with tidymodels, we use : vfold_cv.
Adding to our workflow
We can reuse our previously created workflow, we use fit_resamples (instead of the fit function) for training.
# Resampling results
# 5-fold cross-validation using stratification
# A tibble: 5 × 4
splits id .metrics .notes
<list> <chr> <list> <list>
1 <split [340/86]> Fold1 <tibble [2 × 4]> <tibble [0 × 3]>
2 <split [340/86]> Fold2 <tibble [2 × 4]> <tibble [0 × 3]>
3 <split [341/85]> Fold3 <tibble [2 × 4]> <tibble [0 × 3]>
4 <split [341/85]> Fold4 <tibble [2 × 4]> <tibble [0 × 3]>
5 <split [342/84]> Fold5 <tibble [2 × 4]> <tibble [0 × 3]>Collect CV metrics
# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.831 5 0.00482 Preprocessor1_Model1
2 roc_auc binary 0.913 5 0.00621 Preprocessor1_Model1- 0.831 is cross-validated estimate of accuracy for KNN with
K=6 (as that is what was specified inknn_spec) - It is the average accuracy obtained from the 5 validation sets
Deeper look into CV metrics
| id | .metric | .estimate |
|---|---|---|
| Fold1 | accuracy | 0.8488372 |
| Fold2 | accuracy | 0.8255814 |
| Fold3 | accuracy | 0.8235294 |
| Fold4 | accuracy | 0.8235294 |
| Fold5 | accuracy | 0.8333333 |
\[\begin{equation} \dfrac{0.849 + 0.826 + 0.824 + 0.824 + 0.833 }{5} = 0.831 \end{equation}\]
Comments
The standard error (
std_err) is a measure of how uncertain we are in the mean value.Roughly speaking, we can expect the true average accuracy of the classifier to be somewhere roughly between 83% and 83% (
mean\(\pm\)std_err)You may ignore the other columns in the metrics data frame, as they do not provide any additional insight.
You can also ignore the entire second row with
roc_aucas this metric is beyond the scope of this course.
More folds
cancer_vfold <- vfold_cv(cancer_train, v = 10, strata = Class)
vfold_metrics <- knn_wflow |>
fit_resamples(resamples = cancer_vfold) |>
collect_metrics()
vfold_metrics# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.847 10 0.0131 Preprocessor1_Model1
2 roc_auc binary 0.918 10 0.00686 Preprocessor1_Model1In practice, 5- or 10-fold are popular choices
Parameter value selection
We now want to do these for a range of possible values of K. If we go back to our original model specification, we can make the following tweaks:
The tidymodels package collection provides a very simple syntax for tuning models: each parameter in the model to be tuned should be specified as tune() in the model specification rather than given a particular value
Possible values
While we could conceivable, do all possible values from 1 to \(n\) (the number of observations in our training set), we will see that the optimal choice for K will come much earlier than \(n\) = 426.
Then instead of using fit or fit_resamples, we will use the tune_grid function to fit the model for each value in a range of parameter values.
Perform CV on the grid
knn_results <- workflow() |>
add_recipe(cancer_recipe) |>
add_model(knn_spec) |>
tune_grid(resamples = cancer_vfold, grid = k_vals) |>
collect_metrics()
knn_results# A tibble: 40 × 7
neighbors .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 1 accuracy binary 0.822 10 0.0133 Preprocessor1_Model01
2 1 roc_auc binary 0.812 10 0.0150 Preprocessor1_Model01
3 6 accuracy binary 0.847 10 0.0131 Preprocessor1_Model02
4 6 roc_auc binary 0.918 10 0.00686 Preprocessor1_Model02
5 11 accuracy binary 0.857 10 0.0129 Preprocessor1_Model03
6 11 roc_auc binary 0.928 10 0.00895 Preprocessor1_Model03
7 16 accuracy binary 0.857 10 0.0183 Preprocessor1_Model04
8 16 roc_auc binary 0.929 10 0.00898 Preprocessor1_Model04
9 21 accuracy binary 0.866 10 0.0149 Preprocessor1_Model05
10 21 roc_auc binary 0.929 10 0.00917 Preprocessor1_Model05
# ℹ 30 more rowsAccuracy check
# A tibble: 20 × 7
neighbors .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 1 accuracy binary 0.822 10 0.0133 Preprocessor1_Model01
2 6 accuracy binary 0.847 10 0.0131 Preprocessor1_Model02
3 11 accuracy binary 0.857 10 0.0129 Preprocessor1_Model03
4 16 accuracy binary 0.857 10 0.0183 Preprocessor1_Model04
5 21 accuracy binary 0.866 10 0.0149 Preprocessor1_Model05
6 26 accuracy binary 0.859 10 0.0136 Preprocessor1_Model06
7 31 accuracy binary 0.843 10 0.0152 Preprocessor1_Model07
8 36 accuracy binary 0.848 10 0.0149 Preprocessor1_Model08
9 41 accuracy binary 0.846 10 0.0171 Preprocessor1_Model09
10 46 accuracy binary 0.855 10 0.0160 Preprocessor1_Model10
11 51 accuracy binary 0.850 10 0.0157 Preprocessor1_Model11
12 56 accuracy binary 0.846 10 0.0185 Preprocessor1_Model12
13 61 accuracy binary 0.853 10 0.0136 Preprocessor1_Model13
14 66 accuracy binary 0.853 10 0.0140 Preprocessor1_Model14
15 71 accuracy binary 0.855 10 0.0147 Preprocessor1_Model15
16 76 accuracy binary 0.855 10 0.0147 Preprocessor1_Model16
17 81 accuracy binary 0.846 10 0.0178 Preprocessor1_Model17
18 86 accuracy binary 0.846 10 0.0197 Preprocessor1_Model18
19 91 accuracy binary 0.853 10 0.0174 Preprocessor1_Model19
20 96 accuracy binary 0.852 10 0.0131 Preprocessor1_Model20Plotting accuracy
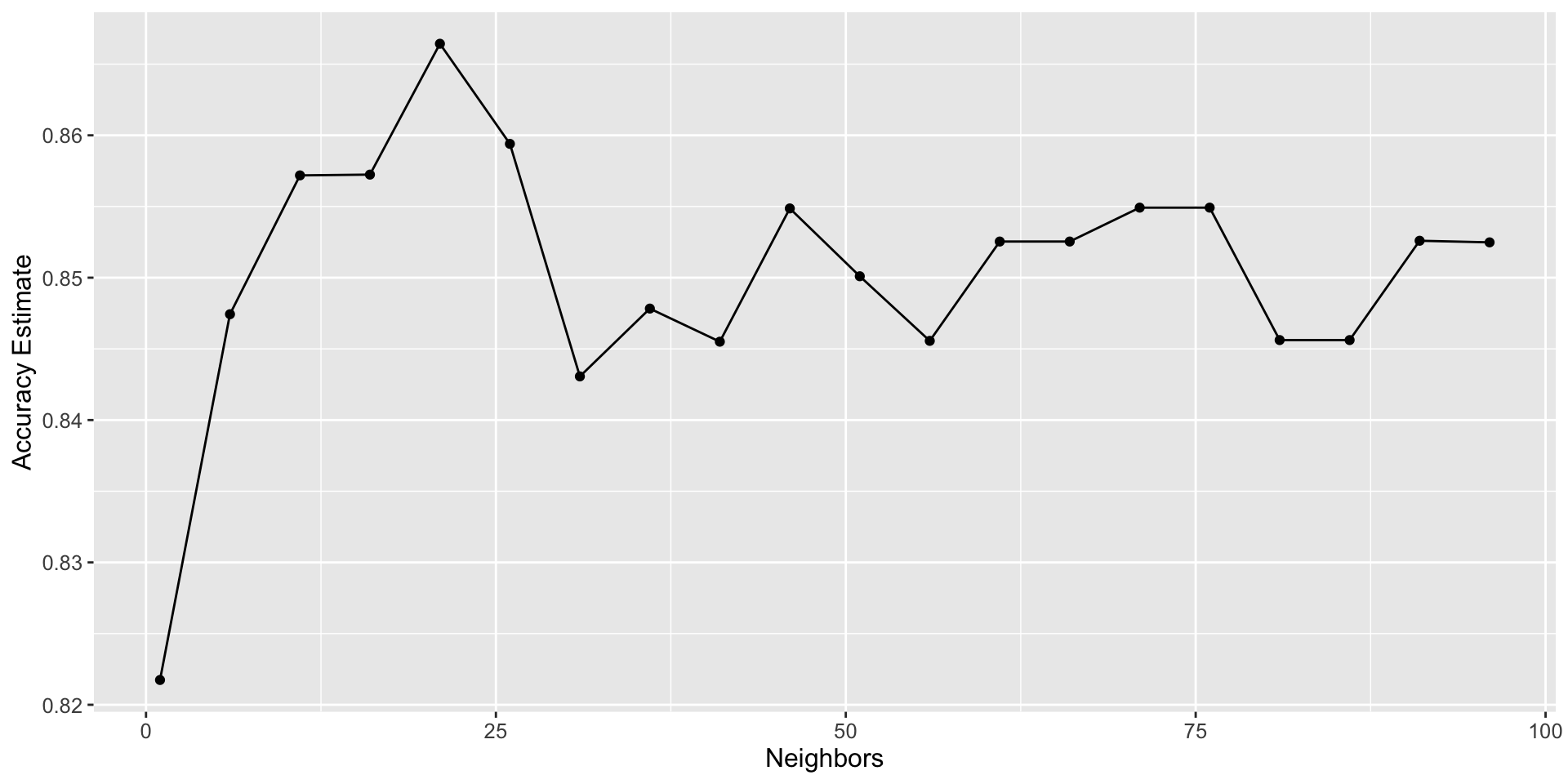
We can decide which number of neighbors is best by plotting the accuracy versus K
Plotting accuracy
Best K?
- Setting the number of nearest neighbours to
K=bestKprovides the highest accuracy (87%) - But there is no exact or perfect answer here;
K= andK= would be reasonably justified, as all of these differ in classifier accuracy by a small amount. - Remember: the values you see on this plot are estimates of the true accuracy of our classifier.
- Although
K=bestKvalue is higher than the others on this plot, that doesn’t mean the classifier is actually more accurate with this parameter value!
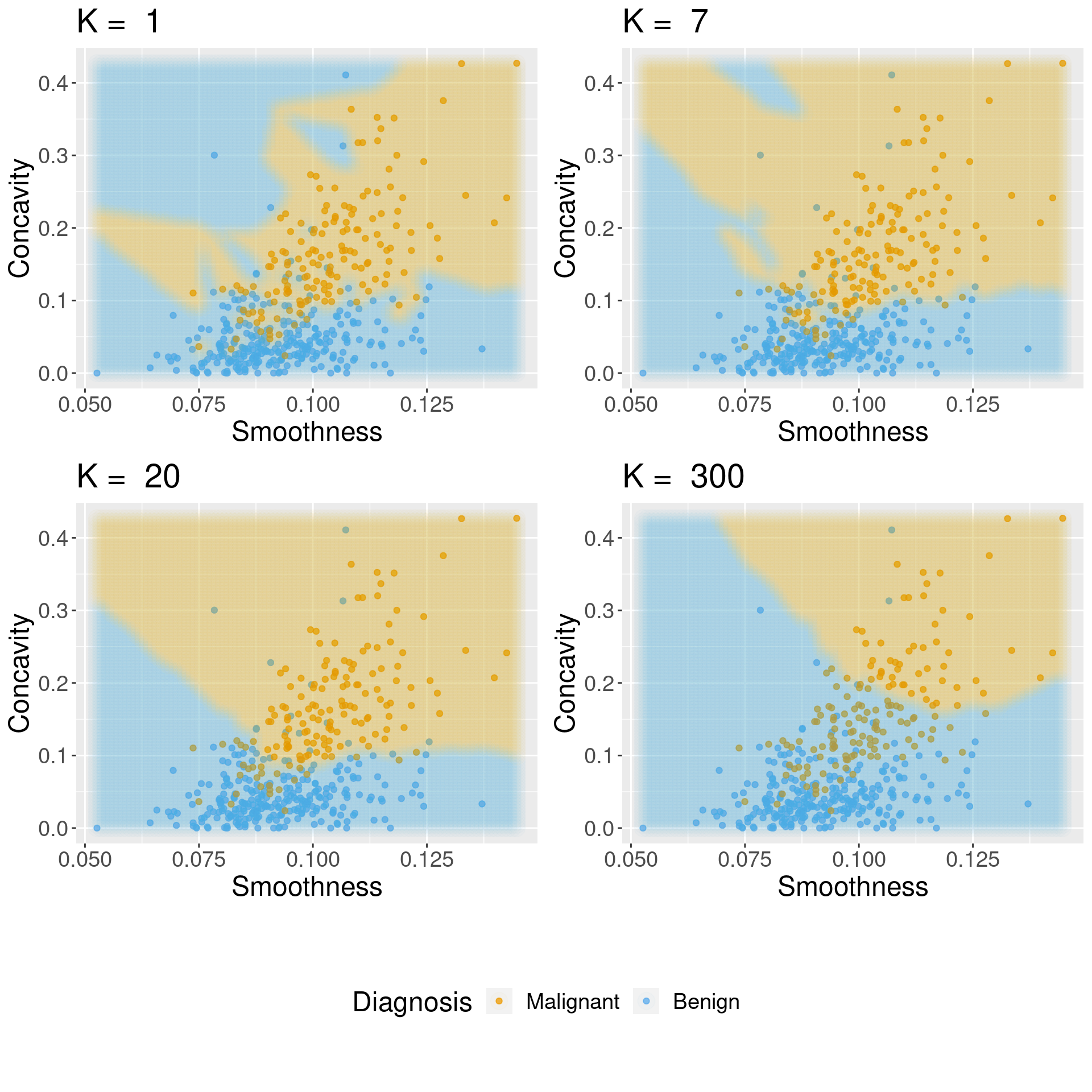
Comment
Generally, when selecting K (and other parameters for other predictive models), we are looking for a value where:
- we get roughly optimal accuracy, so that our model will likely be accurate;
- changing the value to a nearby one (e.g., adding or subtracting a small number) doesn’t decrease accuracy too much, so that our choice is reliable in the presence of uncertainty;
- the cost of training the model is not prohibitive (e.g., in our situation, if
Kis too large, predicting becomes expensive!).
Summary
Comment