# install.packages("devtools")
# devtools::install_github("rstudio/EDAWR")
library(EDAWR)
data(cases)
casesLecture 6: Data Wrangling: Part 2
DATA 101: Making Prediction with Data
University of British Columbia Okanagan
Outline
In today’s lecture we’ll be look
Introduction
- As discussed previously, the process of preparing your data for analysis is often a tedious and long process.
- Therefore any tools and tricks we can obtain to facilitate this process will be worth the investiment of time and energy.
- Last class we started using the dplyr package—a core package from —for managing data.
- Today we will take advantage of another core package from tidyverse called tidyr.
tidyr
The goal of tidyr is to help you create “tidy” data wherein:
- each row is a single observation,
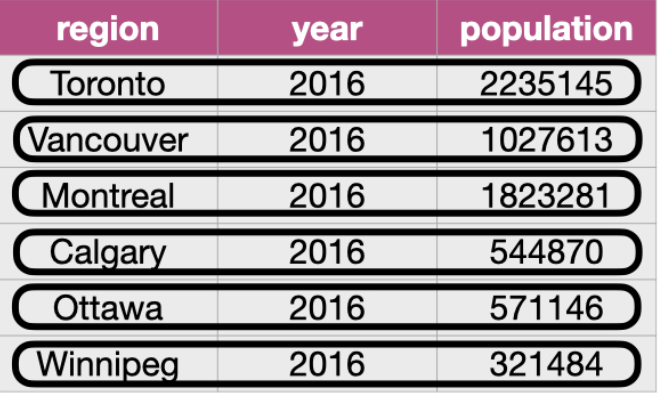
tidyr
The goal of tidyr is to help you create “tidy” data wherein:
- each row is a single observation,
- each variable is a single column, and
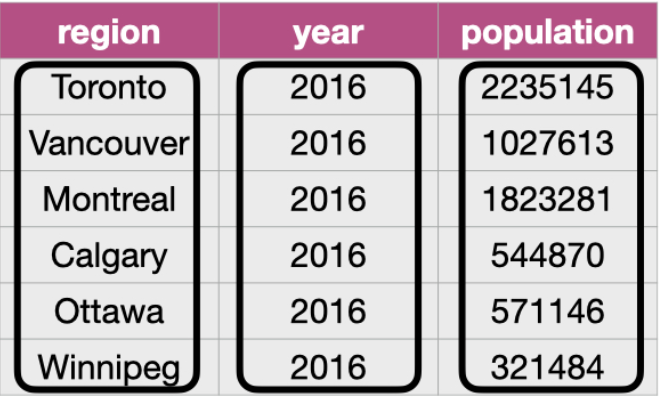
tidyr
The goal of tidyr is to help you create “tidy” data wherein:
- each row is a single observation,
- each variable is a single column, and
- each single cell contains a single value
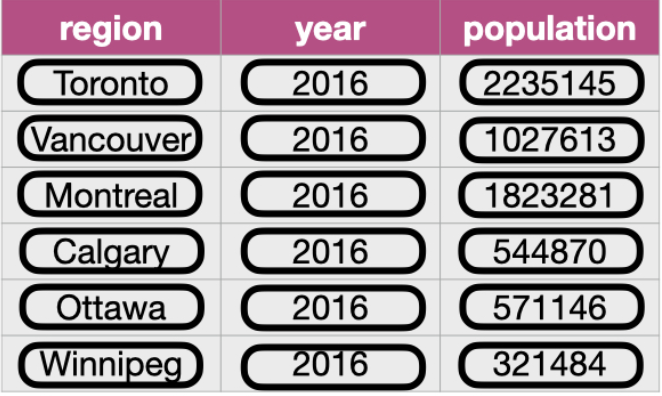
Pivoting Data
“Pivoting” data, in the context of data wrangling, refers to the process of reorganizing or restructuring a dataset from a long format to a wide format or vice versa.
This transformation involves changing the arrangement of the data to make it more suitable for downstream analysis.
There are typically two types of pivoting:
- Pivoting from Long to Wide Data
- Pivoting from Wide to Long Data
Wide data
Wide data format is characterized by having many columns and fewer rows.
This format can make it easy to read and understand the data, especially when when you have data with a small number of observations but a large number of variables.
However, wide data can be less suitable for certain types of analysis and visualization, particularly when you want to perform aggregations, comparisons, or create certain types of plots.
Long data
Long data format is characterized by having fewer columns and more rows. It’s often used to represent data with repeated measures or observations over time or categories.
In long data, variables are typically stacked into a single column, and an additional column is used to indicate the context or category to which each observation belongs.
This format is often preferred for data analysis, modeling, and certain types of visualizations, as it’s more amenable to aggregation and summarization.
Example
| ID | Year | level_1 | level_2 | level_3 |
|---|---|---|---|---|
| 1 | 2010 | 10 | 20 | 30 |
| 2 | 2011 | 15 | 25 | 35 |
| 3 | 2012 | 12 | 22 | 32 |
Going from the left table to the right table is pivoting from wide to long.
| ID | Year | Variable | Value |
|---|---|---|---|
| 1 | 2010 | level_1 | 10 |
| 1 | 2010 | level_2 | 20 |
| 1 | 2010 | level_3 | 30 |
| 2 | 2011 | level_1 | 15 |
| 2 | 2011 | level_2 | 25 |
| 2 | 2011 | level_3 | 35 |
| 3 | 2012 | level_1 | 12 |
| 3 | 2012 | level_2 | 22 |
| 3 | 2012 | level_3 | 32 |
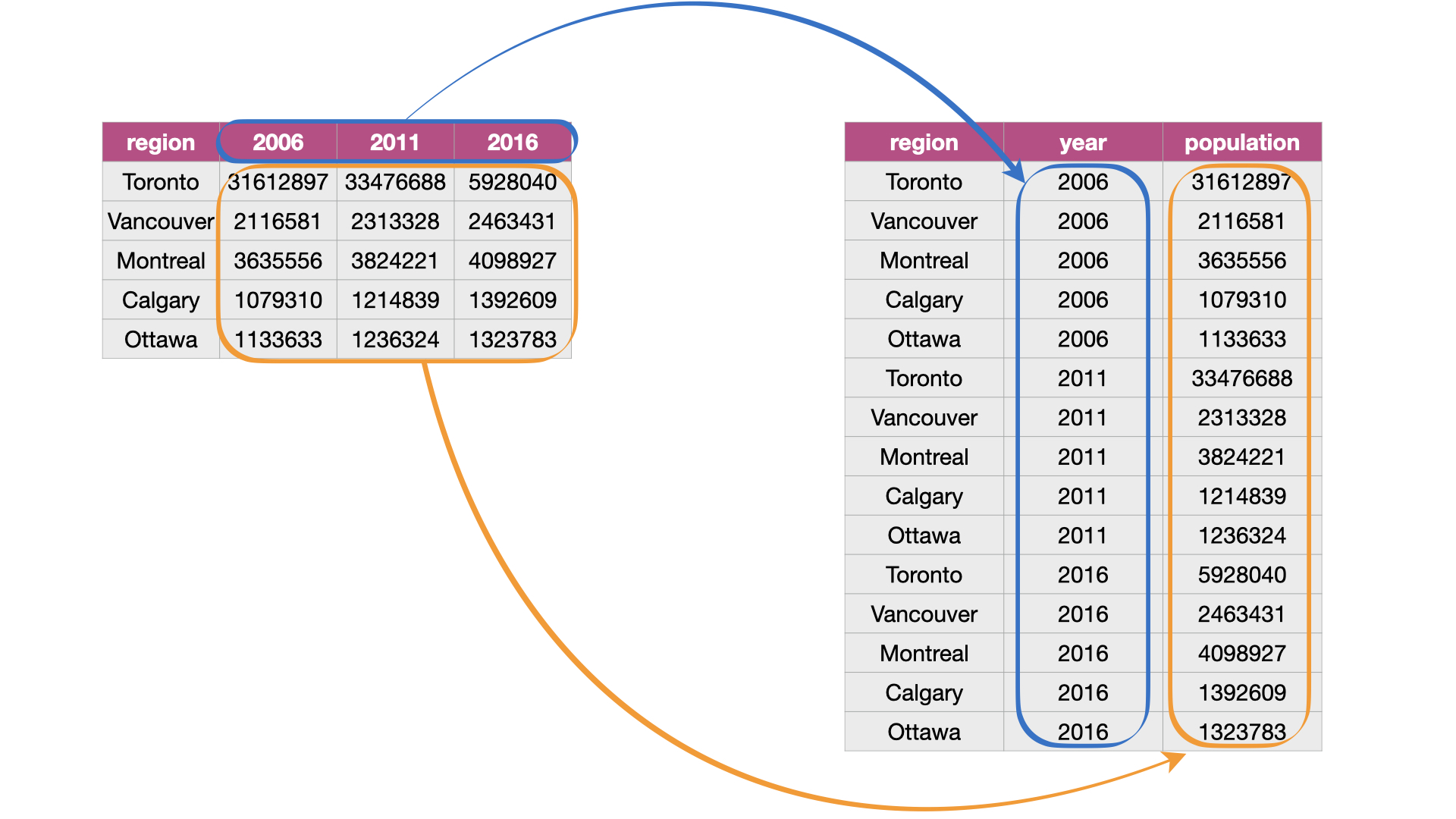
Source: Figure 3.7 from DS book: Pivoting data from a wide to long data format.
Problems the wide table
While the wide table is easy to for humans to read it difficult to work with when performing analysis using R.
- e.g. if we wanted to find the latest year we would have to extract the column names, store them to a vector, coerce to dates, then apply a function like
max().
- e.g. if we wanted to find the latest year we would have to extract the column names, store them to a vector, coerce to dates, then apply a function like
The problem only gets worse if you would like to find the value for the population for a given region for the latest year.
Furthermore, we don’t know what the numbers under each year actually represent.
Functions for pivoting
- It is worth mentioning that there is a modern and outdated way of doing this.
- Both use tidyr functions to transform data between long and wide but the newer functions are recommended.
Old function:
gather()spread()
New functions:
pivot_longer()pivot_wider()
Pivoting rom wide to long
Example: cases
gather
The
gather()function is used to pivot data from a wide format into a long format.This functions work with key-value pairs.
Key: the column names in the original wide dataset that you want to stack or gather into a single column.
Value: the data in the cells corresponding to the key columns.
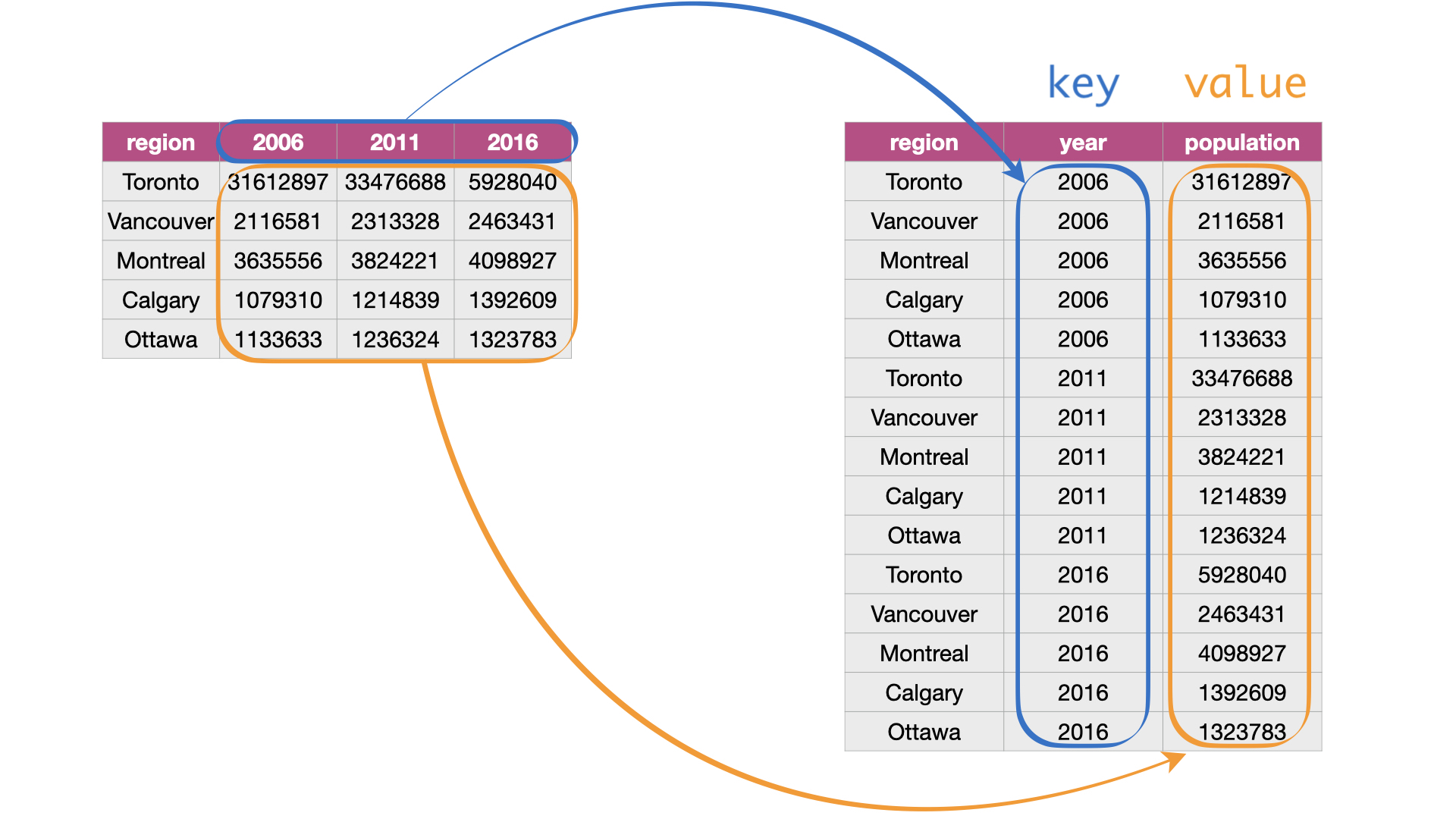
Adapted from Source: Figure 3.7 from DS book
syntax
The general syntax for gather:
dataa data frame you want to cleankeyname of new key column (character string)valuename of the new value column (character string)...a selection of columns to collapse (e.g.2:4)
Warning
If you look at the help file for
gather()will notice that it says that the lifecycle is superseded which is just a softer version of depreciated.A superseded function has a known better alternative, but the function itself is not going away.
A superseded function will not emit a warning (since there’s no risk if you keep using it), but the documentation will tell you what is recommend instead.
Out with the old, in with the new
Development on gather() is complete, and for new code we recommend switching to pivot_longer(), which is easier to use, more featureful, and still under active development.
is equivalent to
pivot_longer
pivot_longer()makes datasets longer by increasing the number of rows and decreasing the number of columns.pivot_longer()is commonly needed to tidy raw datasets as they often optimise for ease of data entry or ease of comparison rather than ease of analysis.The inverse transformation is
pivot_wider()
syntax
dataa data frame to pivot.cols<tidy-select> columns to pivot into longer formatnames_toA character vector specifying the new column or columns to create from the information stored in the column names ofdataspecified bycols.values_toa string specifying the name of the column to create from the data stored in cell values...Additional arguments passed on to methods.
Example revisited
pivot_longer(
1 cases,
2 cols = 2:4,
3 names_to = "year",
4 values_to = "n"
) # output on next slide ...- 1
- data set we want to reshape
- 2
- columns we want to combine
- 3
- “year” the name of the column to be created (the values will come from the names of the columns we want to combine)
- 4
- “n” is the name of the column to be created (the values will come from the values of the columns we want to combine)
Example revisited
Pivoting from long to wide
pivot_wider
Suppose we have observations spread across multiple rows rather than in a single row.
pivot_wider()is the opposite ofpivot_longer(): it makes a dataset wider by increasing the number of columns and decreasing the number of rows.pivot_longer()takes a set of columns and pivots them into two columns: one for variable names one for values.pivot_wider()takes key-value pairs and spreads them into multiple columns based on the unique values in the key column.
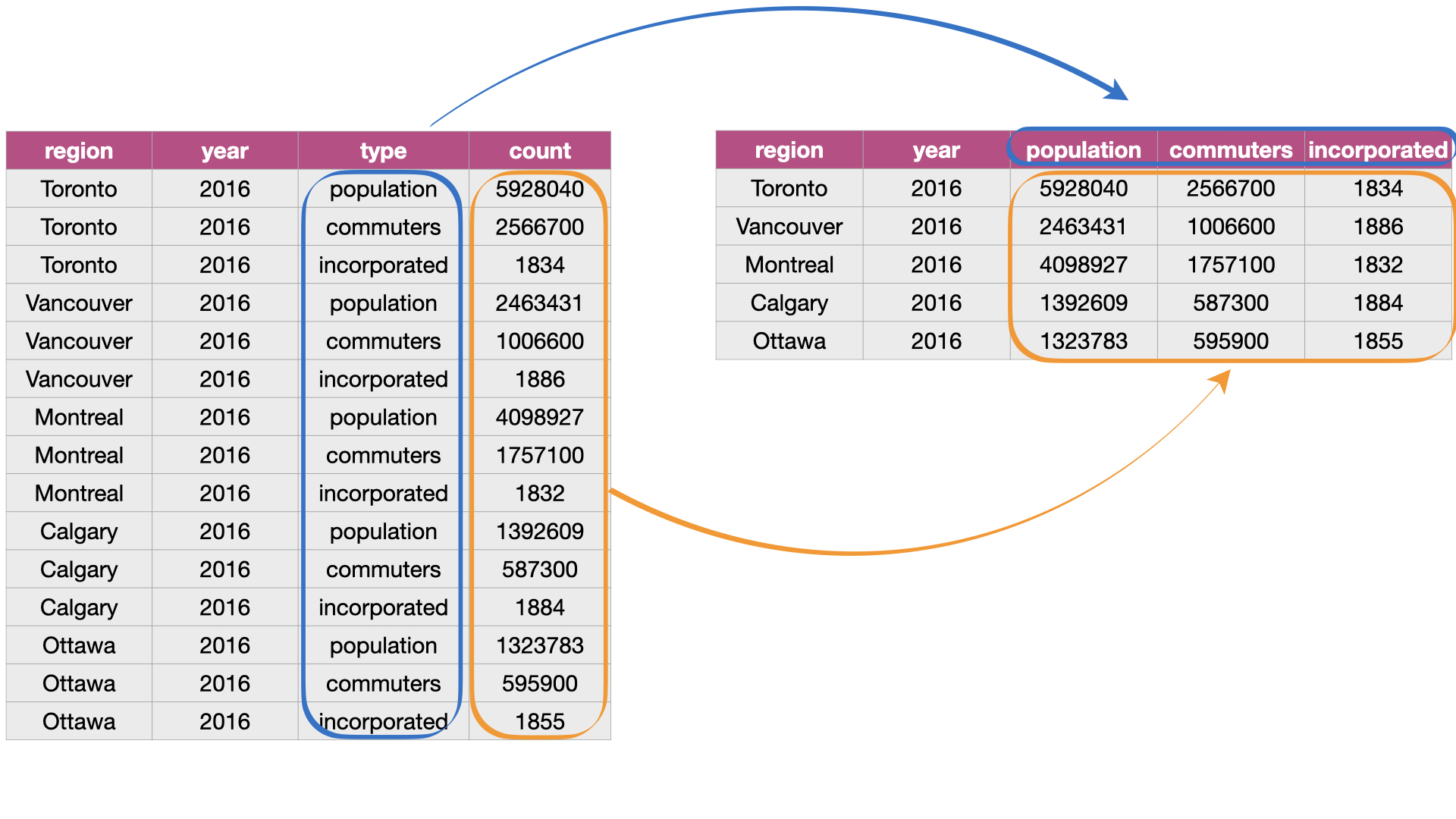
Sourced from Fig 3.10 of DS book
Problems with long table
This data is not “tidy”1
Observation (here, population, commuter, and incorporated values for a region) is split across three rows.
Using data in this format—where two or more variables are mixed together in a single column—makes it harder to apply many usual tidyverse functions.
- e.g. to find the maximum number of commuters would require an additional step of filtering for the commuter values before the maximum can be computed.
syntax
This function generally increases the number of columns (widens) and decreases the number of rows in a data set.
dataa data frame to pivot…additional arguments passed on to methodsnames_from/values_from: <tidy-select> a pair of arguments describing which column(s) to get the name of the output column (names_from), and which column(s) to get the cell values from (values_from).
Example: pollution
Example: pollution
Example: pollution
separate
Another handy tidyr is
separate()It is used to split a single column of data that contains multiple values separated by a delimiter into multiple columns
You specify the delimiter or separator that separates the values within the original column as a regular expression or numeric locations
syntax
This function uses the following basic syntax:
where:
data: Name of the data framecol: Name of the column to separateinto: Vector of names for the column to be separated intosep: The value to separate the column at
Example 1
ex1 <- data.frame(player=c('A', 'A', 'B', 'B', 'C', 'C'),
year=c(1, 2, 1, 2, 1, 2),
stats=c('22-2', '29-3', '18-6', '11-8', '12-5', '19-2'))
ex1The delimiter in this case is the hyphen -
Goal: separate the stats column into two new columns called “points” and “assists” as follows:
Comment
- The data is now tidy; however, we aren’t done yet!
- Notice data type of each column is character (
<chr>). - Because of the delimiter (-) R read these columns in as character types, and by default,separate()will return columns as character data types. - Fortunately, the
separate()function can convert these to the appropriate data type.
convert
Now we can see they are being converted to integers
group_by and summarize
Last lecture we saw how we can combine summarize() and group_by() to summarize values for subgroups within a data set.
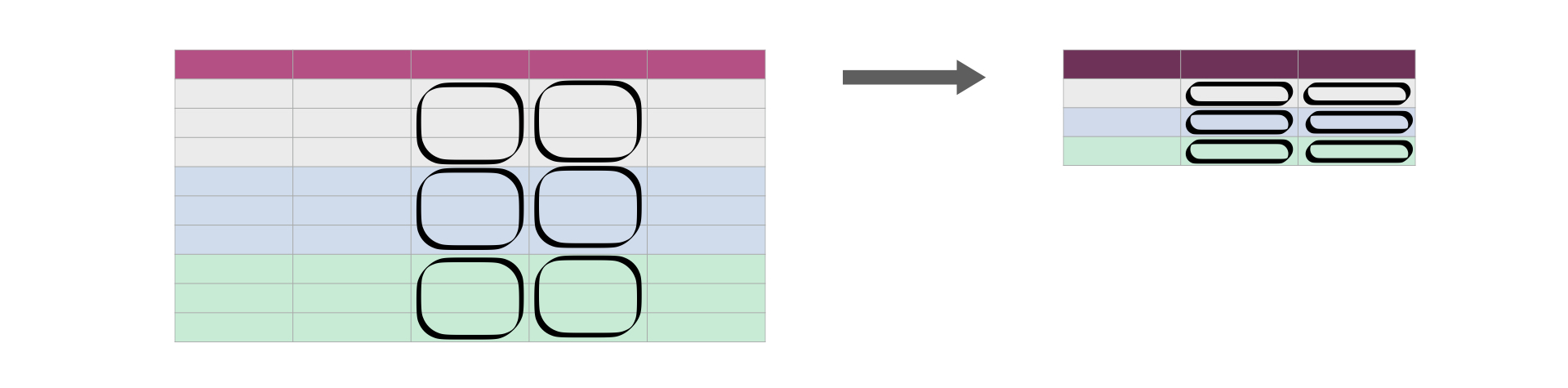
Sourced from Fig 3.16 of DS book
Example: chicago
across and summarize
Now we’ll see how we can use the summarize() function across many columns.
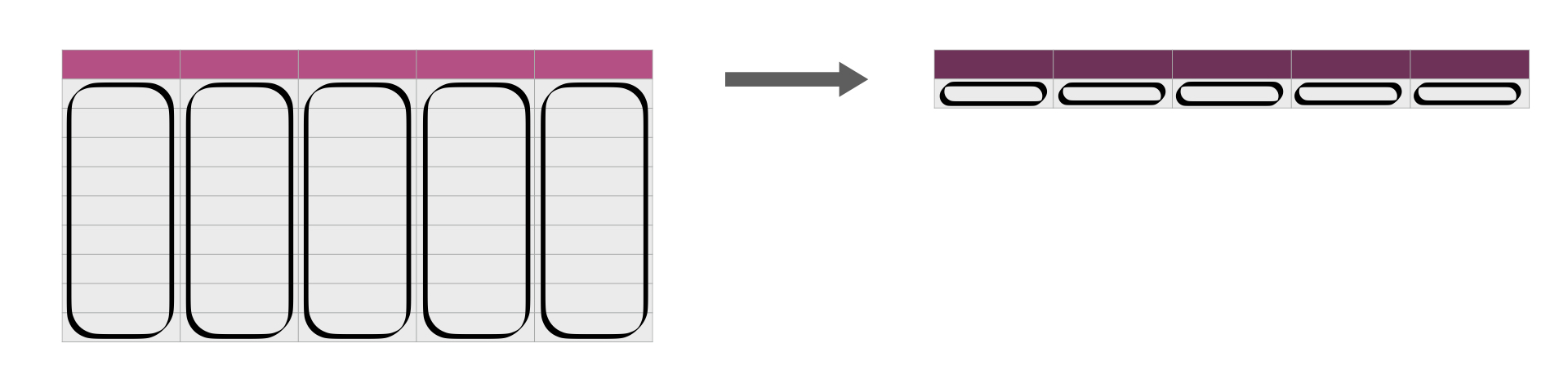
Sourced from Fig 3.17 of DS book
summarize + across
To summarize statistics across many columns, we can use the
summarize()andacross()functions from the dplyr package.The
summarize(across(...))function combination allows you to apply a summary function across multiple columns of a data frame or tibble.To do this more efficiently, we can pair
summarize()withacrossand use a colon:to specify a range of columns we would like to perform the statistical summaries on.
syntax
dfa data frame or tibble.cols<tidy-select> columns to transform.fnsa function name (e.g.meanof a purrr-style lambda function)
- Notice how the
max()summary function returnNAsfor two of the columns - This is because when we apply the
maxsummary function to columns that containNAs - To resolve this issue, again we need to add the argument
na.rm = TRUE
To avoid creating a user-defined function, we could instead create an anonymous function …
Anonymous functions
Anonymous functions, also known as lambda functions or inline functions, are used when you want to define a small, unnamed function for a specific task without formally creating a named function using
function().In purrr, you can create anonymous functions using the tilde (
~) operator.For unary1 functions,
~ .x + 1is equivalent tofunction(.x) .x + 1.
Example revisited
Anonymous function:
map
An alternative to
summarizeandacrossfor applying a function to many columns is themapfamily of functions.Let’s again redo the previous example, but using
mapwith themaxfunction this time.More generally, the
map()function in the purrr package is used for applying a function to each element of a list or vector and returning a new list.
syntax
The general syntax for map() is
.xan object (a vector, data frame or list) that you want to iterate over.fthe function you would like to apply to each element
⚠️ There is no argument to specify which columns to apply the function to; it will simply apply the function to each column (resp. element) of the dataframe (resp. list/vector)
Example re-revisited
map output type
You’ll notice that the output of the
map()function is a list.While we could convert this to a data frame, a simpler alternative is to use a different
map()function; see?map.
map function |
Output |
|---|---|
map |
list |
map_lgl |
logical vector |
map_int |
integer vector |
map_dbl |
double vector |
map_chr |
character vector |
map_dfc |
data frame, combining column-wise |
map_dfr |
data frame, combining row-wise |
Example
This returns a data frame rather than a list which is perhaps more desirable.
Apply across columns
Sometimes we need to apply a function to many columns in a data frame.
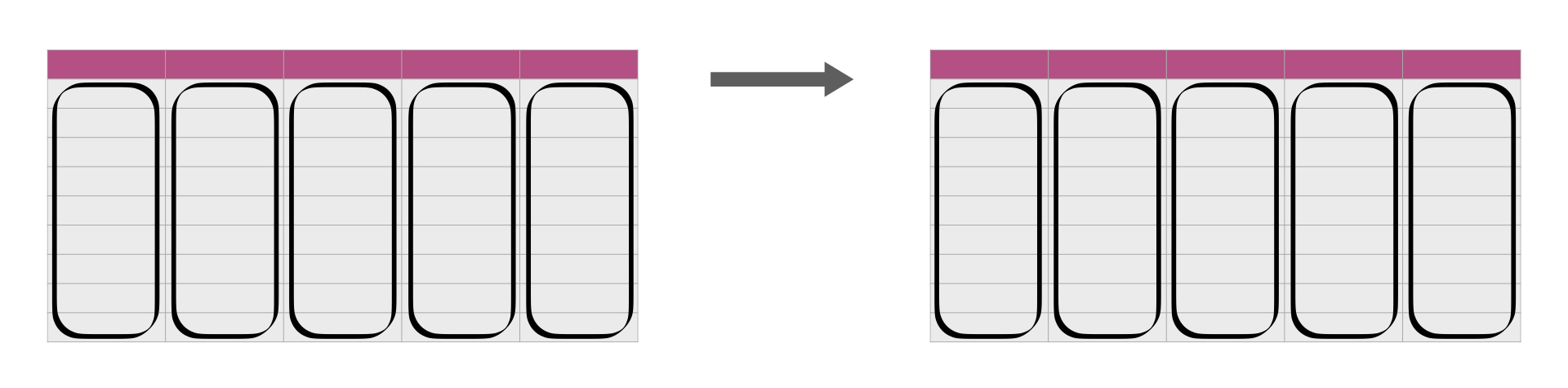
Sourced from Fig 3.18 of DS book
Example
Suppose we want to scale multiple columns (say the pm25tmean2 to the no2tmean2 column) from the chicago data set.
To accomplish such a task, we can use mutate paired with across.
Apply across rows
Sometimes we need to apply a function across columns but within one row:
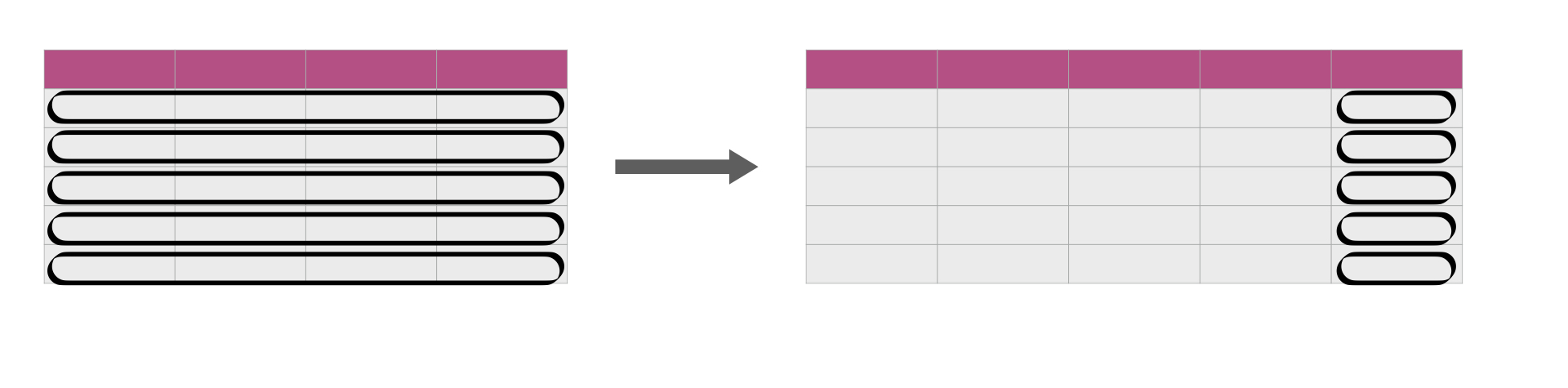
Sourced from Fig 3.19 from DS book
Example
For instance, suppose we want to know the maximum value between pm25tmean2, pm10tmean2, o3tmean2 and no2tmean2 for each record in the chicago data set.
Example
apply
In addition to these tidyverse functions we also have a handy base R function called
apply()apply()is primarily used for applying a function to the rows or columns of a matrix or array. It is not designed for use with lists or other data structures.You specify whether you want to apply the function to rows (MARGIN = 1), columns (MARGIN = 2), or both (MARGIN = c(1, 2)).
syntax
Xan array, including a matrix.MARGINa vector giving the subscripts which the function will be applied over. E.g., for a matrix 1 indicates rows, 2 indicates columns, c(1, 2) indicates rows and columns. Where X has named dimnames, it can be a character vector selecting dimension names.FUNthe function to be applied....optional arguments to FUN.simplifya logical indicating whether results should be simplified if possible.
A simply example
Summary
| Function | Description |
across() |
allows you to apply function(s) to multiple columns |
filter() |
subsets rows of a data frame |
group_by() |
allows you to apply function(s) to groups of rows |
mutate() |
adds or modifies columns in a data frame |
map() |
general iteration function |
pivot_longer() |
makes the data frame longer and narrower |
pivot_wider() |
makes a data frame wider and decreases the number of rows |
rowwise() |
applies functions across columns within one row |
separate() |
splits up a character column into multiple columns |
select() |
subsets columns of a data frame |
summarize() |
calculates summaries of inputs |